Zoysiagrass
Breeding for cold tolerance:
 Our Zoysiagrass germplasm collection has been evaluated for cold tolerance at two locations in the mountains of North Carolina. Lines with cold tolerance were used as parents for development of improved materials. A set of 40 lines that have been selected for their superior cold tolerance, turf quality and faster establishment are currently under evaluation in field trials at multiple locations to test their performance against commercial checks in order to determine their suitability for release.
Our Zoysiagrass germplasm collection has been evaluated for cold tolerance at two locations in the mountains of North Carolina. Lines with cold tolerance were used as parents for development of improved materials. A set of 40 lines that have been selected for their superior cold tolerance, turf quality and faster establishment are currently under evaluation in field trials at multiple locations to test their performance against commercial checks in order to determine their suitability for release.
Understanding levels of speciation within Zoysia spp.:
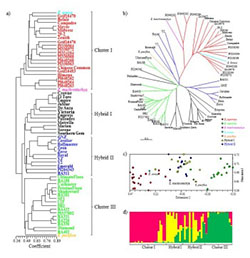 Understanding the population structure present within Zoysia germplasm can assist plant breeders in exploiting available variation. The objectives of this study were to assess simple sequence repeat (SSR) allelic diversity within and among Zoysia spp., evaluate the genetic constitution of putative interspecific hybrids, and determine if Zoysia spp. and hybrids can be differentiated by inflorescence traits. Based on findings, this study was able to verify the presence of Z. japonica x Z. matrella hybrids. The sheer number of hybrids identified speaks to both the historical and future applicability of hybrid-based breeding strategies within zoysiagrass. Admixture present between Zoysia accessions further validates the hypothesis that Zoysia spp. are actually subpopulations or ecotypes within one species and not separate species.
Understanding the population structure present within Zoysia germplasm can assist plant breeders in exploiting available variation. The objectives of this study were to assess simple sequence repeat (SSR) allelic diversity within and among Zoysia spp., evaluate the genetic constitution of putative interspecific hybrids, and determine if Zoysia spp. and hybrids can be differentiated by inflorescence traits. Based on findings, this study was able to verify the presence of Z. japonica x Z. matrella hybrids. The sheer number of hybrids identified speaks to both the historical and future applicability of hybrid-based breeding strategies within zoysiagrass. Admixture present between Zoysia accessions further validates the hypothesis that Zoysia spp. are actually subpopulations or ecotypes within one species and not separate species.
Mapping QTL for cold acclimation and freeze tolerance
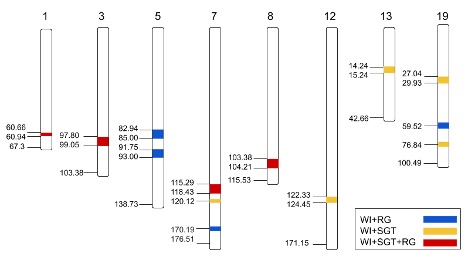
Molecular markers associated with freezing tolerance would be useful for effective selection of winter hardy germplasm before field testing. A pseudo-F2 mapping population of 175 individuals was developed from crosses between Z. japonica cultivars Meyer (freeze-tolerant) and Victoria (freeze-susceptible) and used to generate a high-density genetic map of 104 SSR markers and 2359 sequencing-derived SNP markers. The map covers 324 Mbp and 2520 cM as well as the 20 chromosomes for the zoysiagrass haploid genome.
For field evaluation, phenotypic data on winter injury, establishment, and turf quality collected in North Carolina and Indiana in 2014–2016 were used in conjunction with linkage map to identify quantitative trait loci (QTL) associated with winter hardiness. Fifty-six QTL associated with winter injury, establishment, and turf quality were identified over six environments. Seven regions of interest were identified on chromosomes 8, 11, and 13 where colocation of QTL for three or more traits occurred.
Cold acclimation and freezing tests were investgated in controlled environment for post-freeze Surviving Green Tissue (SGT) and Regrowth (RG) evaluation. Forty QTL regions were identified for SGT, and forty-one QTL regions for RG, accounting for 6.4-12.2% of the phenotypic variation (R2). Thirteen regions of interest overlapped with putative winter injury QTL identified in the above field study. Overlapping QTL between the winter injury and controlled freezing studies were compared to putative proteins associated with cold acclimation, where five expressed protein families were observed to be present in the annotated genes, including ATPase family proteins, NADP superfamily proteins, protein kinase superfamily proteins, and auxin-responsive family proteins. Following further validation, these QTL and their associated markers may be utilized in future breeding efforts for the development of a broader pool of zoysiagrass cultivars capable of surviving in cool-humid climates.
Transcriptomic analysis of freeze response
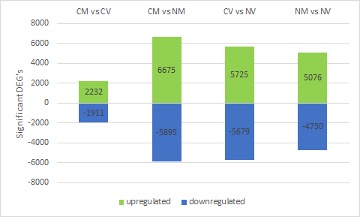
To deepen our understanding of the physiologic and metabolic components of freezing tolerance in zoysiagrass, a transcriptomic approach was utilized to identify genes involved in cold acclimation, a critical step in the development of freezing tolerance. Meyer, a freeze tolerant cultivar, and Victoria, a freeze susceptible cultivar were used to identify changes in expression in freeze tolerance associated transcripts. Differentially expressed genes and their involved pathways of interest were identified. Several candidate genes demonstrated significant fold change differences between treatments and cultivars, including genes encoding low molecular mass early light-inducible proteins, late embryogenesis abundant proteins, auxin-responsive proteins, and photosystem II proteins. Further investigation of these genes and their function may contribute to increase our understanding of molecular mechanism in freezing tolerance.
Proteomics analysis of cold acclimation
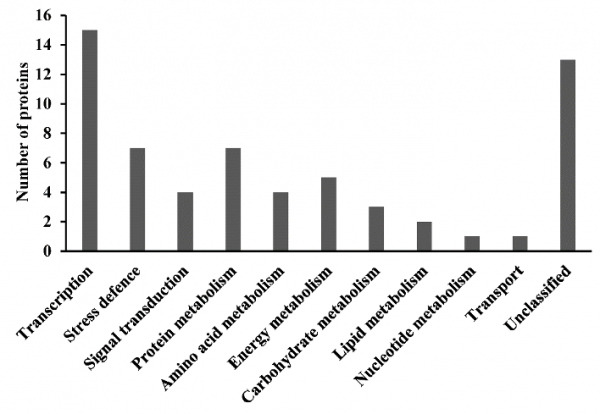
We investigated the relationship between cold acclimation, protein expression, and freeze tolerance in cold-acclimated (CA) and non-acclimated (NCA) plants of Zoysia japonica Steud. cultivars ‘Meyer’ (freeze-tolerant) and ‘Victoria’ (freeze-susceptible). Overall, proteomic analysis identified 62 protein spots differentially accumulated in abundance under cold acclimation. Nine and 22 unique protein spots were identified for Meyer and Victoria, respectively, with increased abundance or decreased abundance. In addition, 23 shared protein spots were found among the two cultivars in response to cold acclimation. Function classification revealed that these proteins were involved primarily in transcription, signal transduction and stress defense, carbohydrate and energy metabolism, protein and amino acid metabolism. Several proteins of interest for their association with cold acclimation were identified. Further investigation of these proteins and their functional categories may contribute to increase our understanding of the differences in freezing tolerance among zoysiagrass germplasm.
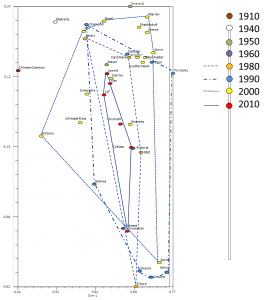
Assessing changes in genetic diversity of Zoysiagrass commercial cultivars released from 1910 through 2016:
We’re analyzing levels of allelic diversity at the gene and population levels in Zoysia cultivars released between 1910 and 2016. Simple sequence repeat (SSR) markers are being used to determine whether allelic diversity over has changed among these cultivars. This study will help elucidate the influence a century of science-based plant breeding has had in zoysiagrass breeding. Knowing these effects will help maintain or renew efforts to increase genetic diversity in this economically important turfgrass species.