Centipedegrass
Mutation breeding in centipedegrass:
 The goal of this project is to generate morphological and stress tolerance variation through the use of a chemical mutagen. During 2013 and 2015, a total of 4,600 M1 progenies were planted as nurseries in Sandhills Research Station and the Upper Mountain Research Station in Laurel Springs. Nurseries were under evaluation for aggressiveness, genetic color, and leaf texture. Based on performance, 85 lines were selected and compared to five commercial lines in replicated trials at three locations. Evaluated traits include aggressiveness, genetic color, leaf texture and density, uniformity, drought and tolerance, flowering density and seed production rate. Currently, 23 superior lines have been selected to explore the potential of centipedegrass as a seeded crop.
The goal of this project is to generate morphological and stress tolerance variation through the use of a chemical mutagen. During 2013 and 2015, a total of 4,600 M1 progenies were planted as nurseries in Sandhills Research Station and the Upper Mountain Research Station in Laurel Springs. Nurseries were under evaluation for aggressiveness, genetic color, and leaf texture. Based on performance, 85 lines were selected and compared to five commercial lines in replicated trials at three locations. Evaluated traits include aggressiveness, genetic color, leaf texture and density, uniformity, drought and tolerance, flowering density and seed production rate. Currently, 23 superior lines have been selected to explore the potential of centipedegrass as a seeded crop.
Chromosome doubling using colchicine

It has been found that plant structures increase size by duplicating the set of chromosome in the cells. We are working on using colchicine in centipedegrass to develop artificial lines with morphological variations and increase resistance against biotic stresses such as pests and pathogens (insects, nematodes, virus, and diseases). Centipedegrass seeds will be treated with different concentrations of colchicine at different exposure times. Seedlings recovered after treatment will be screened for genome size changes using flow cytometry and later corroborated through chromosome counts. Lines will be planted in the field for further evaluation of turf quality performances.
NGS and SSR development in Centipedegrass
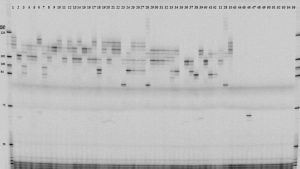 Centipedegrass (Eremochloa ophiuroides) is a warm-season grass well adapted to Southeastern United States. Although it is known for requiring less inputs and presenting higher tolerance to pests, it is also known for the narrow genetic base of the available cultivars. Next-generation sequencing data is being used to develop simple sequence repeat (SSR) markers for centipedegrass. The initial objective of this project is to assess genetic diversity in a collection of accessions collected in central and southern China in the late 90’s. In the future, we expect to use these SSRs to perform marker-assisted selection, analyze population structure, and identify unknown varieties.
Centipedegrass (Eremochloa ophiuroides) is a warm-season grass well adapted to Southeastern United States. Although it is known for requiring less inputs and presenting higher tolerance to pests, it is also known for the narrow genetic base of the available cultivars. Next-generation sequencing data is being used to develop simple sequence repeat (SSR) markers for centipedegrass. The initial objective of this project is to assess genetic diversity in a collection of accessions collected in central and southern China in the late 90’s. In the future, we expect to use these SSRs to perform marker-assisted selection, analyze population structure, and identify unknown varieties.